Pushing the Boundaries of Proteomics Exploration
High throughput proteomics is revolutionizing biomedical research by bridging the gap between genomics and our understanding of human diseases. This breakthrough technology enables researchers to unlock the secrets of the human proteome that were previously not possible. The ability to simultaneously measure thousands of proteins in plasma has opened the doors to identifying previously undiscovered biomarkers, expediting drug discovery, and facilitating large-scale population studies to explore complex diseases and their underlying mechanisms. Using a multi-omics approach, researchers can create a comprehensive map of the molecular pathways that drive the development, progression, and outcomes of diseases by connecting gene expression profiles and genetic variations to protein functions. This valuable insight into the relationships between gene and protein-level information is transforming the way we predict patient responses, stratify populations, and develop novel therapeutic strategies.
Next-Generation Proteomics with Olink® Explore HT
At the forefront of high throughput proteomics technologies is the new Olink® Explore HT, which pushes the boundaries of proteomics exploration by enabling researchers to accurately measure over 5,400 proteins using only 2 µl of sample. A streamlined, automated workflow provides extraordinary scalability for high-throughput data acquisition to help researchers extract actionable biological insights faster. Olink® Explore HT delivers over 80% more protein coverage than the previous generation product, with a 4-fold faster workflow and 6-fold reduction in reagents. The innovative dual-recognition, DNA-coupled Proximity Extension Assay (PEA) technology ensures high specificity, minimizing the risk of wrong targets, data misinterpretation, and resource waste.

Realizing the Full Potential of Population-Scale Proteogenomic Studies
The integration of human genomics and proteomics in large population-scale studies is rapidly gaining traction thanks to these technological advancements. Olink® technology recently played a pivotal role in one of the largest global proteomics health studies. The Pharma Proteomics Project, funded by a consortium of 13 leading biopharmaceutical companies, used the Olink® Explore platform to execute proteomic profiling of blood plasma from over 54,000 participants within the UK Biobank. The analysis covered 3,000 circulating proteins, including many that were previously challenging to capture, and explored their connections to cardiometabolic function, inflammation, neurology, oncology, and COVID-19 outcomes. This Herculean team effort has resulted in the publication of three landmark Nature articles.
Tapping into the Potential of an Open-Access Proteogenomics Atlas
The first Nature paper by Sun et al.1 provides a detailed summary of the dataset, which was used to construct an updated genetic atlas of the plasma proteome. This was achieved through genome wide association study (GWAS)-based proteogenomic analysis and protein quantitative trait loci (pQTL) mapping of 2,923 proteins.
Impressively, this analysis unveiled 14,287 primary genetic associations, with an overwhelming majority (85%) being newly discovered. Numerous associations were uncovered between blood proteins and demographic factors, such as BMI, liver, and kidney functions, and the top 20 most prevalent health conditions. For example, inflammatory cytokines like CXCL17, long thought to contribute towards mental health conditions, were significantly higher in patients with depression. These insights lay the groundwork for a deeper understanding of the molecular mechanisms underlying these health conditions and potentially the subsequent development of more precise diagnostic and therapeutic strategies.
The availability of this open-access proteomics resource is treasure trove for the global research community. It will serve as a vital foundation to help elucidate the biological mechanisms underpinning proteo-genomic discoveries and accelerating the development of biomarkers, predictive models, and therapeutics in the months and years to come1.
The Importance of Analyzing Rare Genetic Variants
AstraZeneca-led research published in the second, companion article2 used the dataset to study the influence of rare genetic variants on the plasma proteome, which has largely remained unexplored.
Using an exome-wide association study (ExWAS) approach, 5,433 rare genotype–protein associations were identified in the Pharma Proteomics Project dataset. Most of these associations (81%) had not been detected in previous GWAS-based studies conducted on the same cohort, highlighting the importance of exome sequencing for rare variant associations.

The authors used variant- and gene-level association tests to map pQTLs across the spectrum of allele frequencies and highlight how these associations can be used in various applications. From the discovery of potential biomarkers associated with a genetic variant linked to fatty liver disease within the HSD17B13 gene to providing insight into the allelic series NLRC4 associated with plasma level changes of the proinflammatory cytokine IL-18, these findings emphasize the significant role of rare pQTLs on biological outcomes. This study also highlights the importance of large-scale proteogenomics studies to uncover associations that offer insights into drug discovery, safety assessments, and drug repurposing applications.
Comparing Proteomics Platforms: Insights from a Comparative Analysis
In the final article3 from deCODE Genetics/Agene, researchers conducted a comparative analysis using proteogenomic data from the Olink® PPP dataset and data obtained from a cohort of 36,000 Icelandic individuals using the SomaScan aptamer-based platform. A subset of 1,514 samples from the Icelandic cohort was also measured using the Olink® Explore 3072 platform to facilitate a direct comparison of protein levels. The researchers utilized genomic data to identify associations between genetic variants and protein levels (pQTLs) and examined both protein levels and pQTLs in relation to available phenotypic data to evaluate performance.
Both studies revealed a substantial number of genetic associations with protein levels, and in this study, the Olink® assays on average exhibited greater precision when compared to the SomaScan based on the lower median coefficient of variation (CV) ratios. The study found that 72% of all proteins measured using Olink® had associated cis-pQTLs compared to 43% while using an aptamer-based technology. The detection of cis-pQTLs provides strong genetic evidence of target specificity, or that the correct target protein is being measured.
The comparative analysis sheds light on the strengths and limitations of high-throughput proteomics platforms. While the study showed that Olink® assays generally exhibited greater precision and genetic validation through cis-pQTLs, it’s important to note that the performance of these platforms may be influenced by the biochemical properties of the assays, especially when quantifying proteins in complex samples like plasma. These orthogonal assays can work complementarily to facilitate population-scale studies to expand the coverage and depth of population-scale proteomic investigations.
A New Era of Multi-Omics
These groundbreaking publications mark an important inflection point in proteomics, where cutting-edge technological advancements like Olink,® Explore are revolutionizing the field, allowing comprehensive and quantitative analysis of the proteome at an unprecedented scale. Its availability heralds a new era of multi-omics research by enabling comprehensive, and quantitative analysis of the proteome at an unprecedented scale. The availability of high-throughput proteomics technology is ushering in a new era of multi-omics research by providing a critical missing piece of the puzzle. This perspective offers insights that were once beyond reach, with the potential to reshape the future of healthcare and precision medicine.
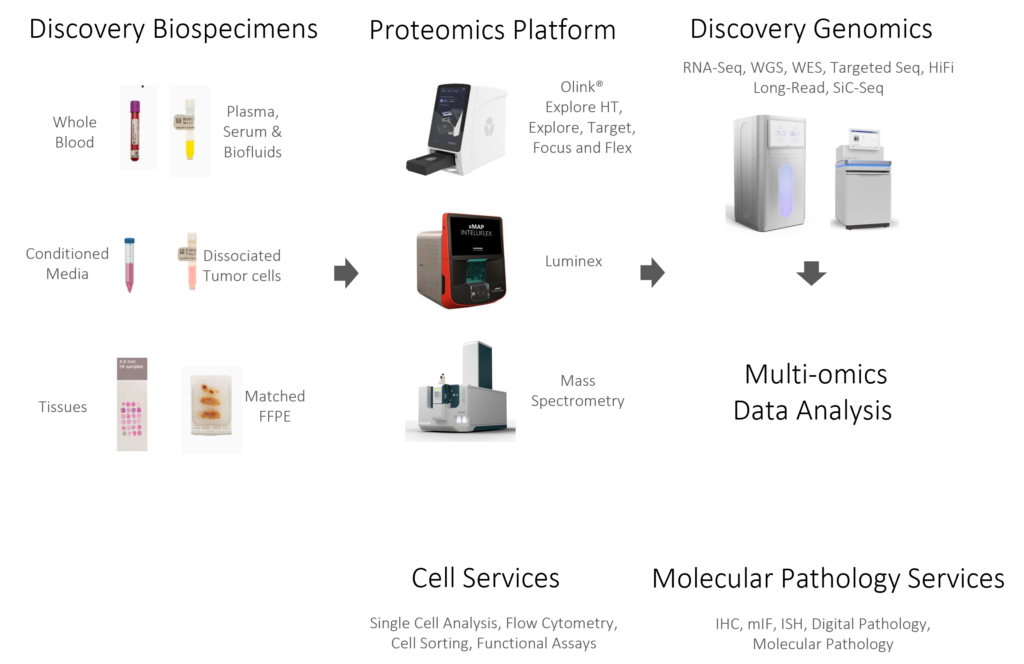
Partnering with Discovery Life Sciences
With Discovery Life Sciences, you have the ideal partner to navigate the exciting frontier of proteomics-driven research. We fuse comprehensive genomics and proteomics services, featuring automated workflows, advanced technologies, and top-tier scientific consultation, with the world’s largest biospecimen inventory and procurement network to provide a singular suite of solutions that accelerates your research journey.
Discovery’s Proteome Center is a certified service provider for Olink Explore HT, Explore, Target, Focus, and Flex platforms as well as other proteomics services using LC-MS/MS, Seer Proteograph MS, and more. Discovery plugs our Proteome Center’s deep expertise and extensive proteomics services into the expansive genomics expertise and NGS-based services of Discovery Genomics. Discovery Genomics has the industry’s largest commercial fleet of NovaSeq X Plus sequencers and large-scale capacity services based on Illumina, PacBio, 10X, and other cutting-edge platforms to support projects of any scale – including population studies.
References
- Sun BB, Chiou J, Traylor M, et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023;622(7982):329-338. doi:10.1038/s41586-023-06592-6
- Dhindsa RS, Burren OS, Sun BB, et al. Rare variant associations with plasma protein levels in the UK Biobank. Nature. 2023;622(7982):339-347. doi:10.1038/s41586-023-06547-x
- Eldjarn GH, Ferkingstad E, Lund SH, et al. Large-scale plasma proteomics comparisons through genetics and disease associations. Nature. 2023;622(7982):348-358. doi:10.1038/s41586-023-06563-x


